This is the full text of a talk I presented at the Parker on the Web 2.0 Symposium in Cambridge on March 16, 2018 (Please note addendum at the end which addresses an issue that came up in discussion later in the day.)
Continue reading “Using VisColl to Visualize Parker on the Web: Reports on an experiment”Dot’s Twitter Bots
I made some Twitter bots! It was mostly very easy.
Continue reading “Dot’s Twitter Bots”How to download images using IIIF manifests, Part II: Hacking the Vatican
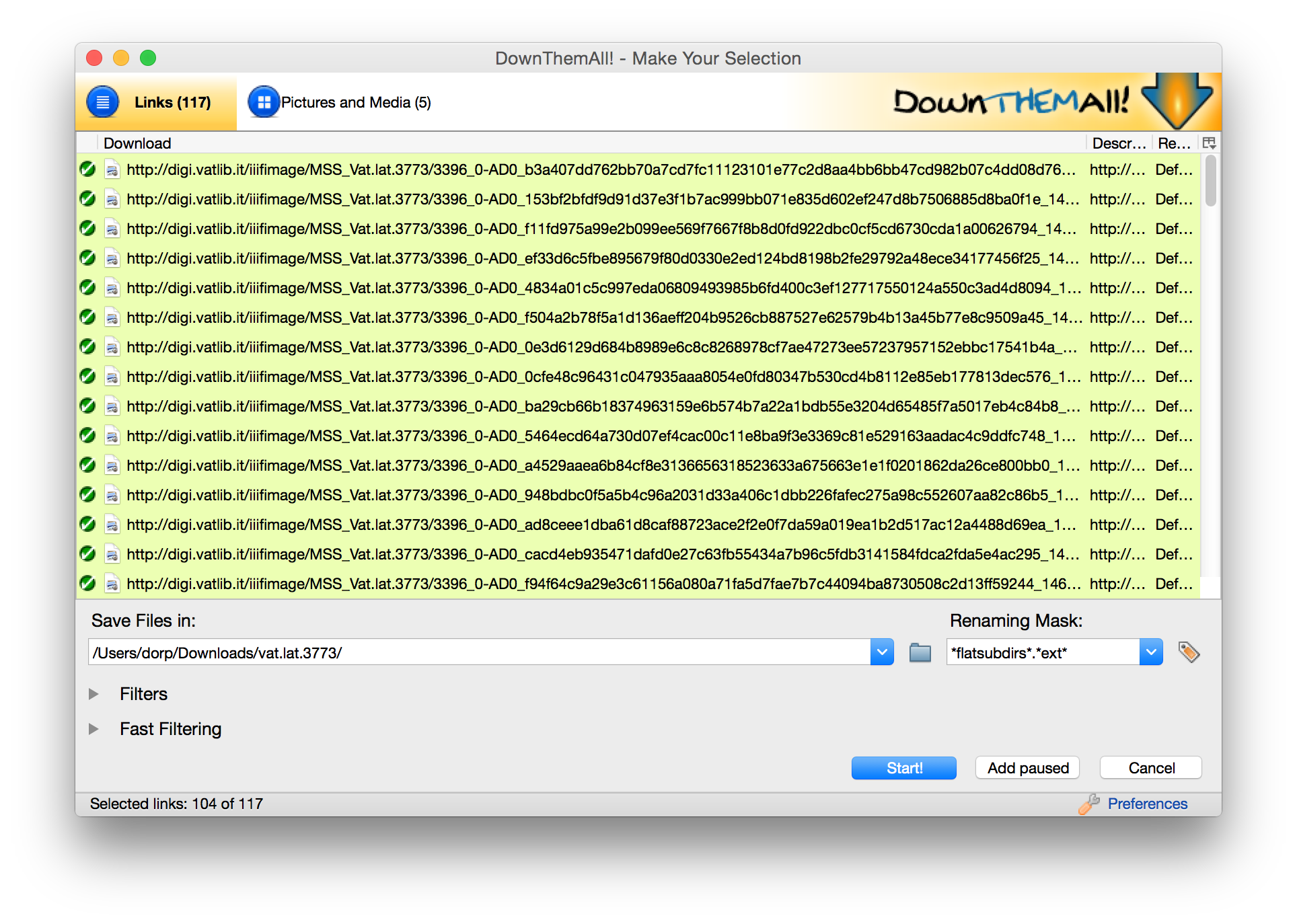
Last week I posted on how to use a Firefox plugin called Down them All to download all the files from an e-codices IIIF manifest (there’s also a tutorial video on YouTube, one of a small but growing collection that will soon include a video outlining the process described here), but not all manifests include direct links to images. The manifests published by the Vatican Digital Library are a good example of this. The URLs in manifests don’t link directly to images; you need to add criteria at the end of the URLs to hit the images. What can you do in that case? In that case, what you need to do it build a list of urls pointing to images, then you can use Down Them All (or other tools) to download them.
Continue reading “How to download images using IIIF manifests, Part II: Hacking the Vatican”How to download images using IIIF manifests, Part I: DownThemAll
IIIF manifests are great, but what if you want to work with digital images outside of a IIIF interface? There are a few different ways I’ve figured out that I can use IIIF manifests to download all the images from a manuscript. The exact approach will vary since different institutions construct their image URLs in different ways. Here’s the first approach, which is fairly straightforward and uses e-codices as an example. Tomorrow I’ll post a second post using on the Vatican Digital Library. Please remember that most institutions license their images, so don’t repost or publish images unless the institution specifically allows this in their license.
Continue reading “How to download images using IIIF manifests, Part I: DownThemAll”UPenn’s Schoenberg Manuscripts, now in PDF
Hi everyone! It’s been almost a year since my last blog post (in which I promised to post more frequently, haha) so I guess it’s time for another one. I actually have something pretty interesting to report!
Continue reading “UPenn’s Schoenberg Manuscripts, now in PDF”